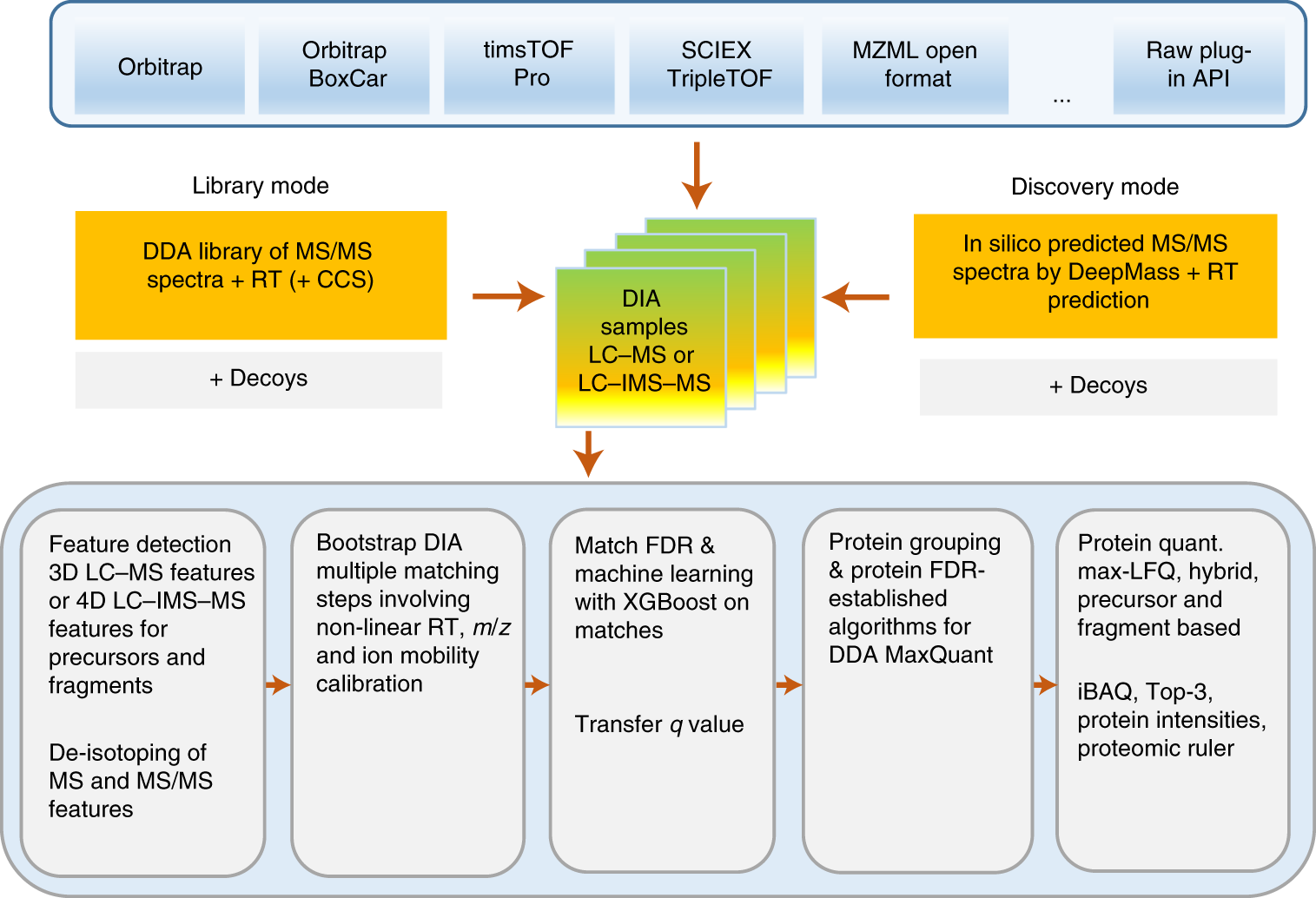
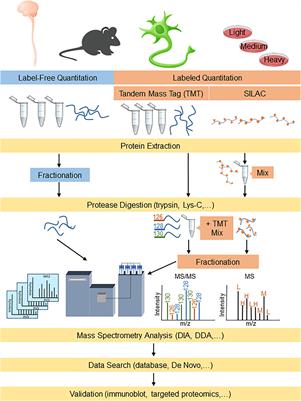
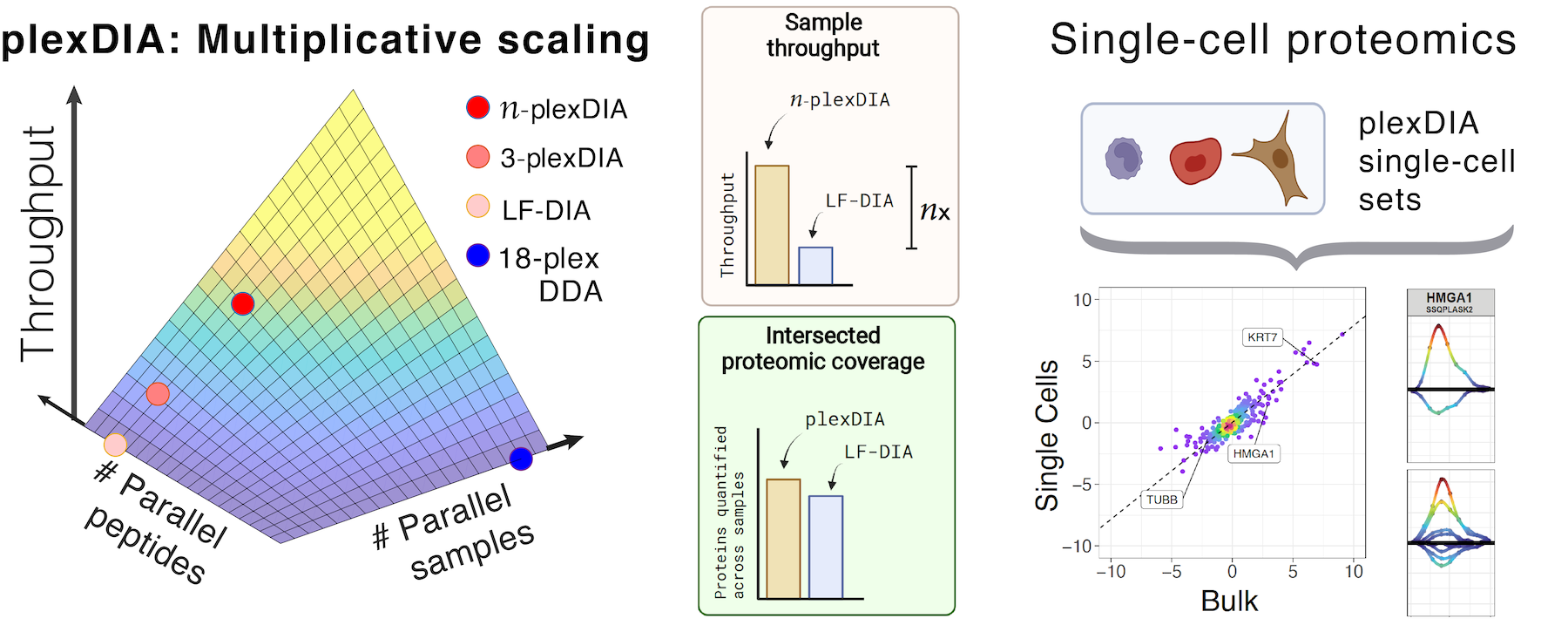
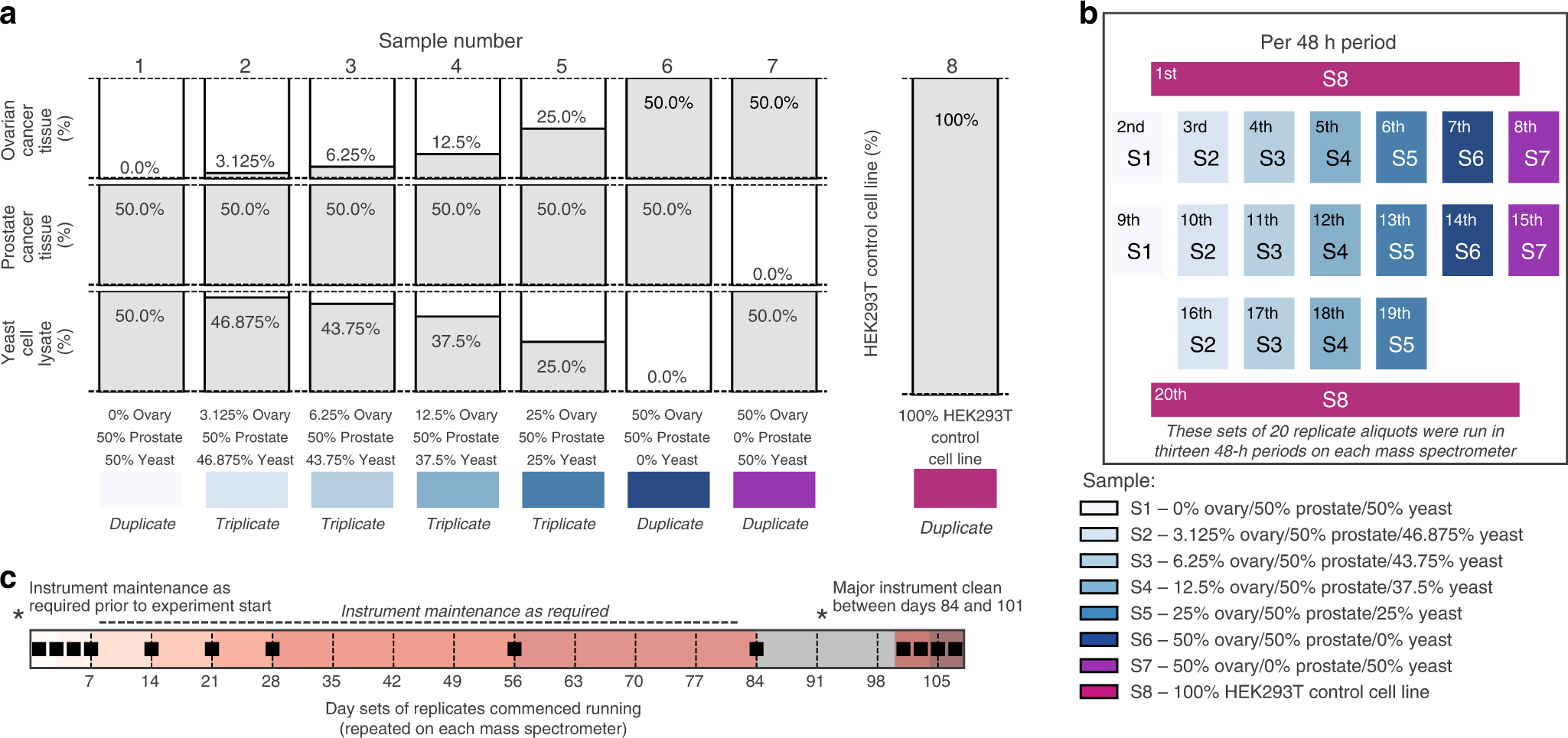
Technical comparison of DDA and different types of DIA in a biological... | Download Scientific Diagram
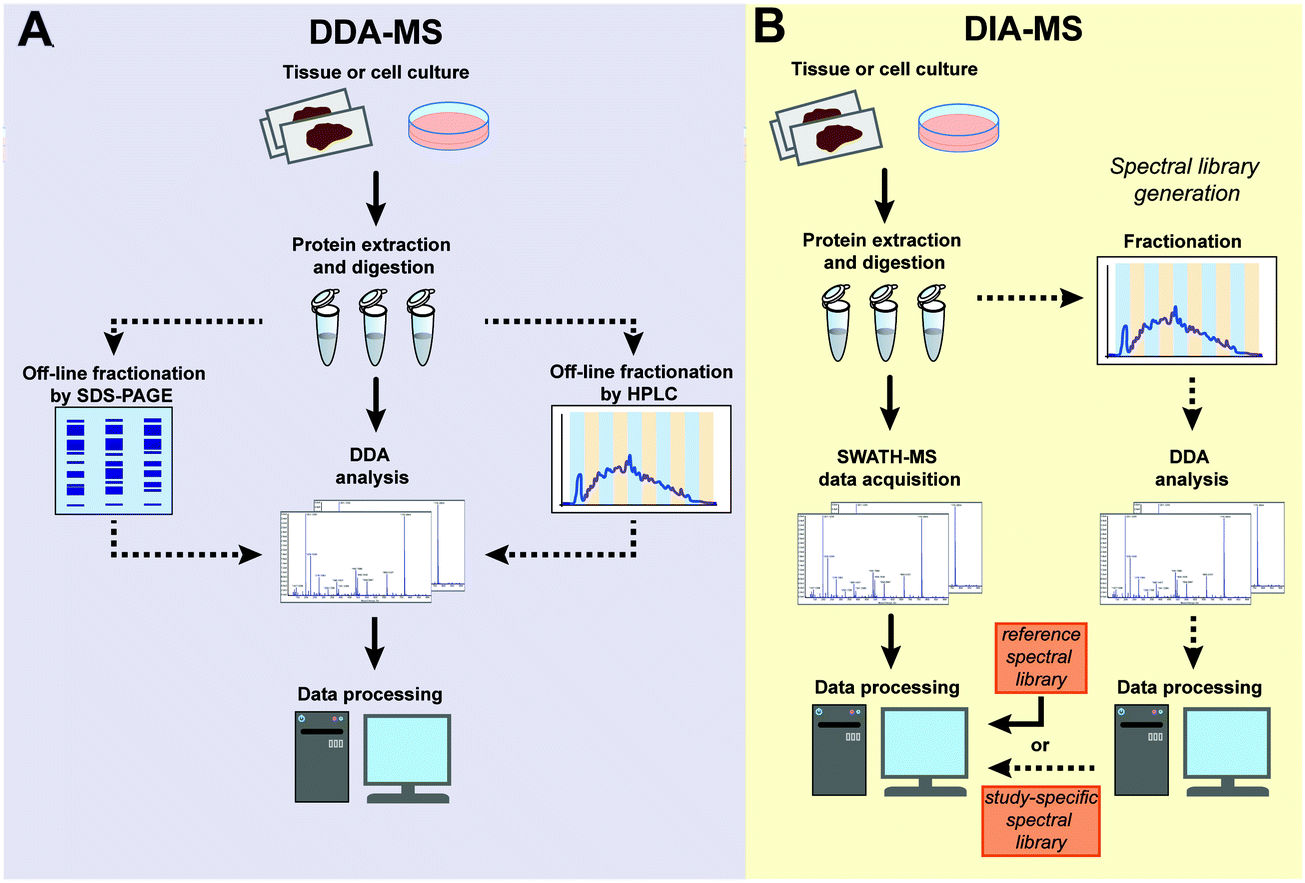
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H
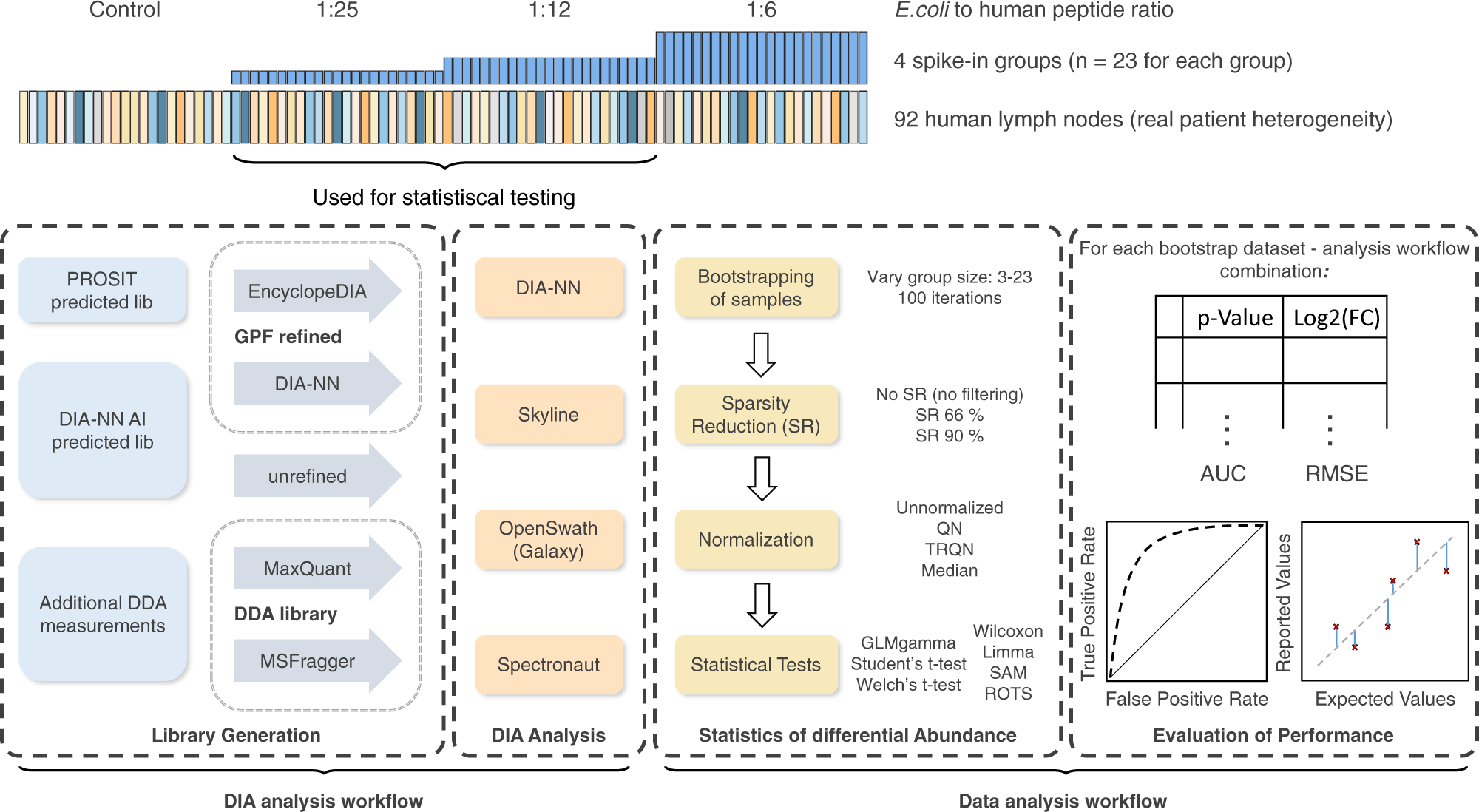
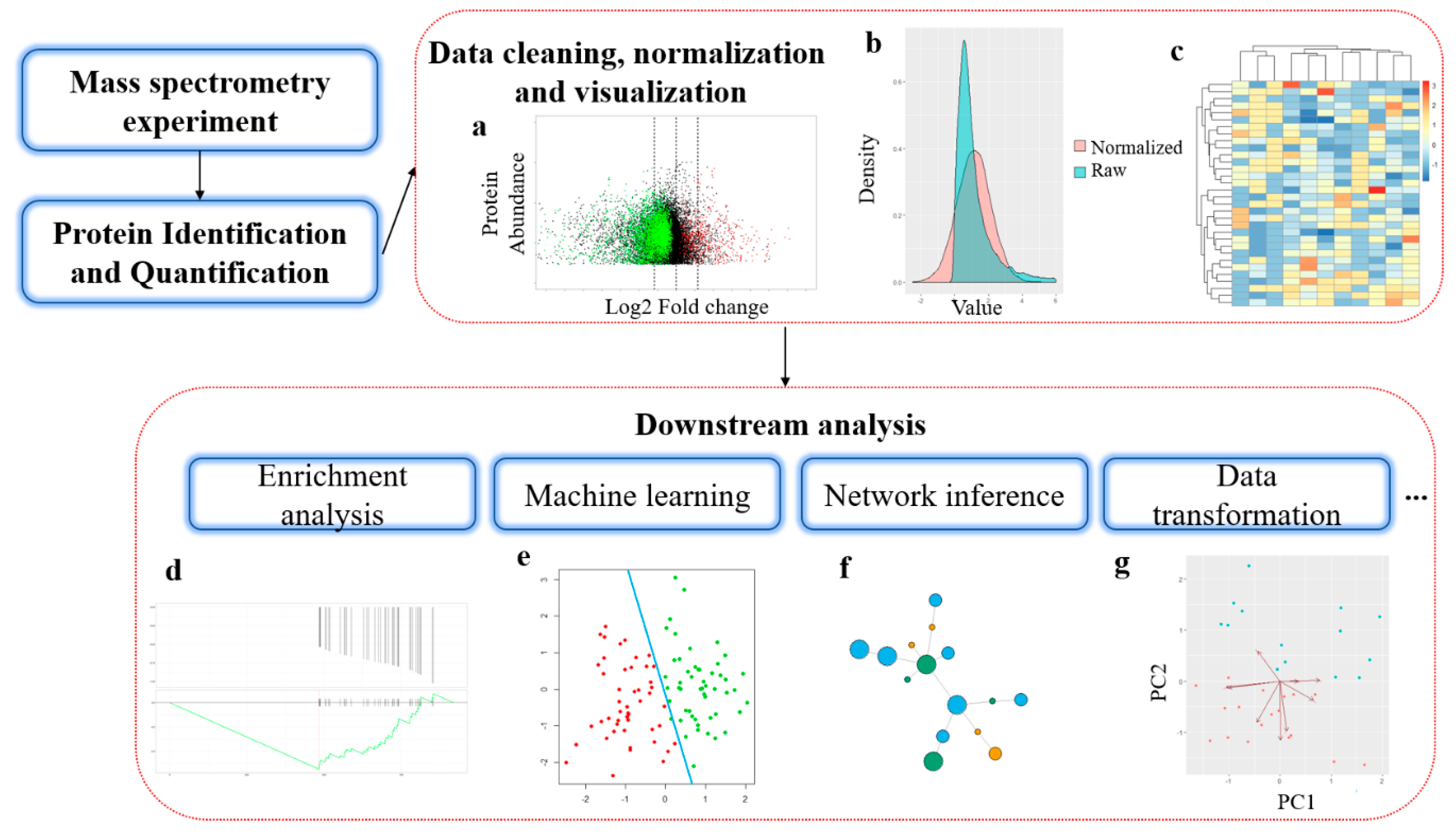
Benchmarking of analysis strategies for data-independent acquisition proteomics using a large-scale dataset comprising inter-patient heterogeneity | Nature Communications
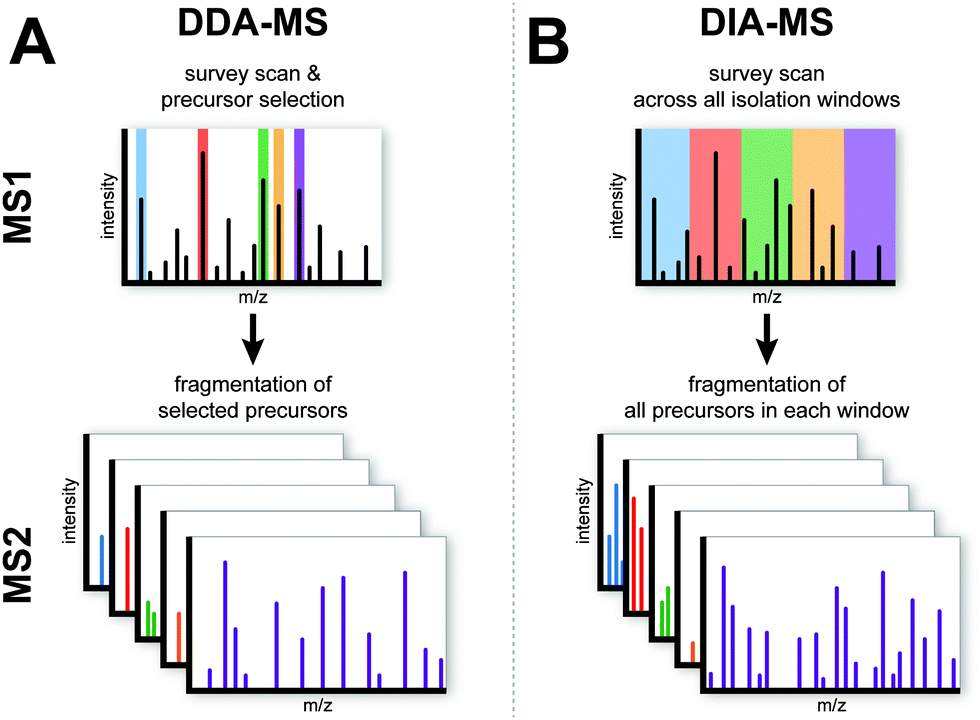
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H
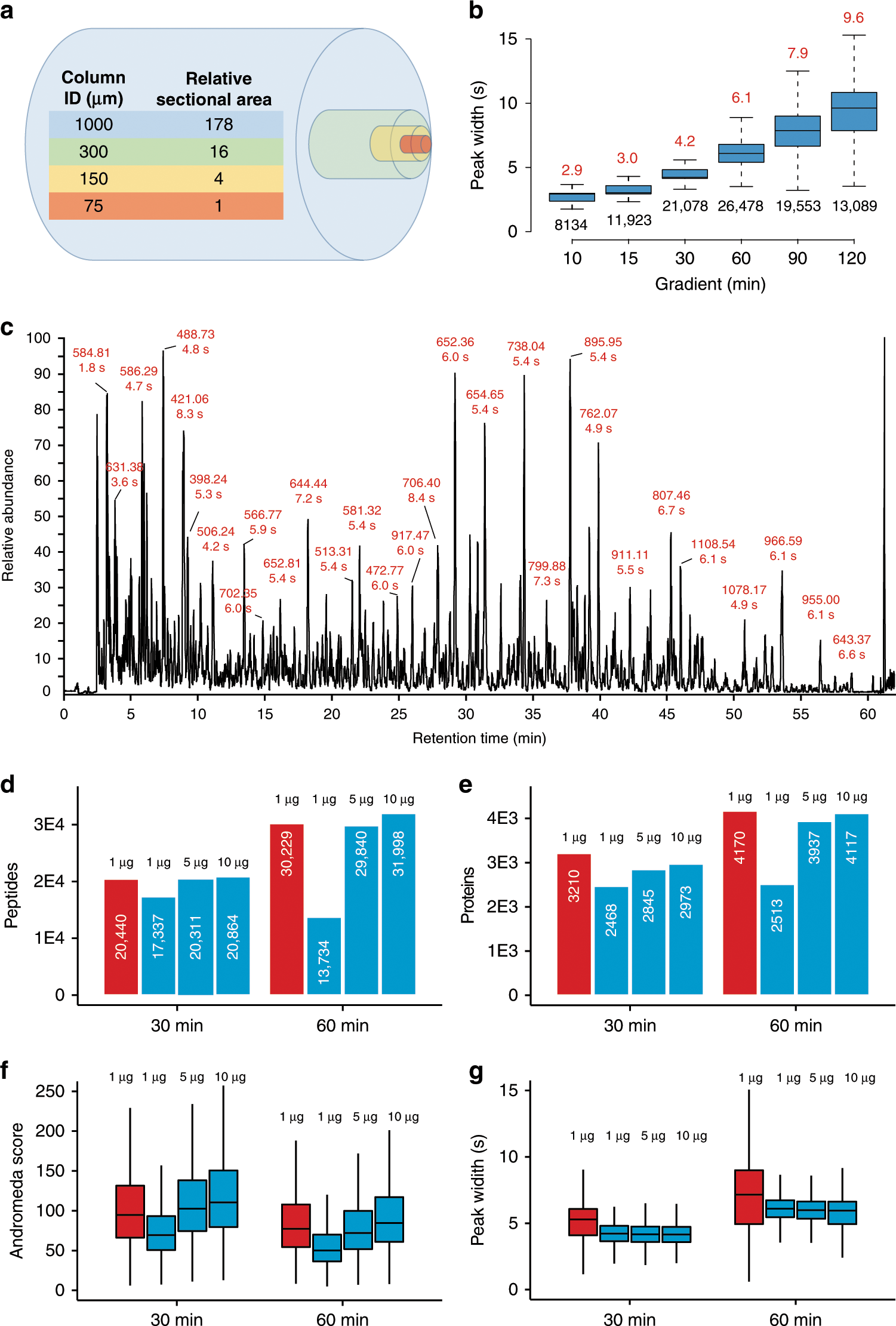
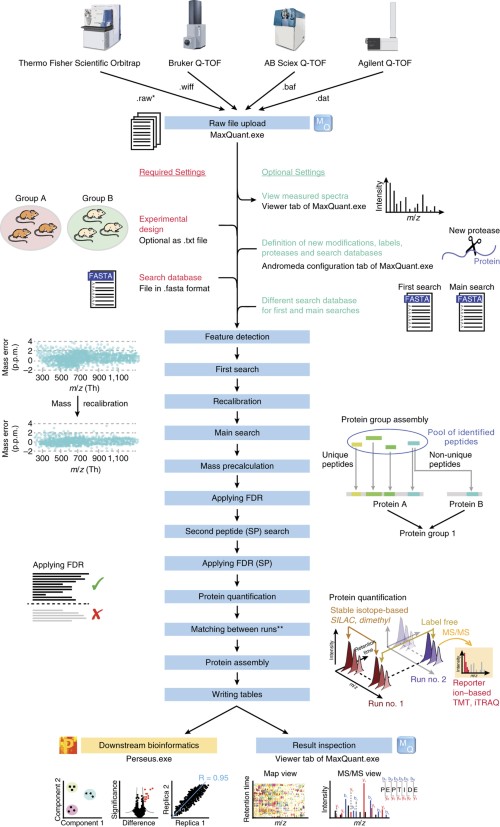
Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS | Nature Communications
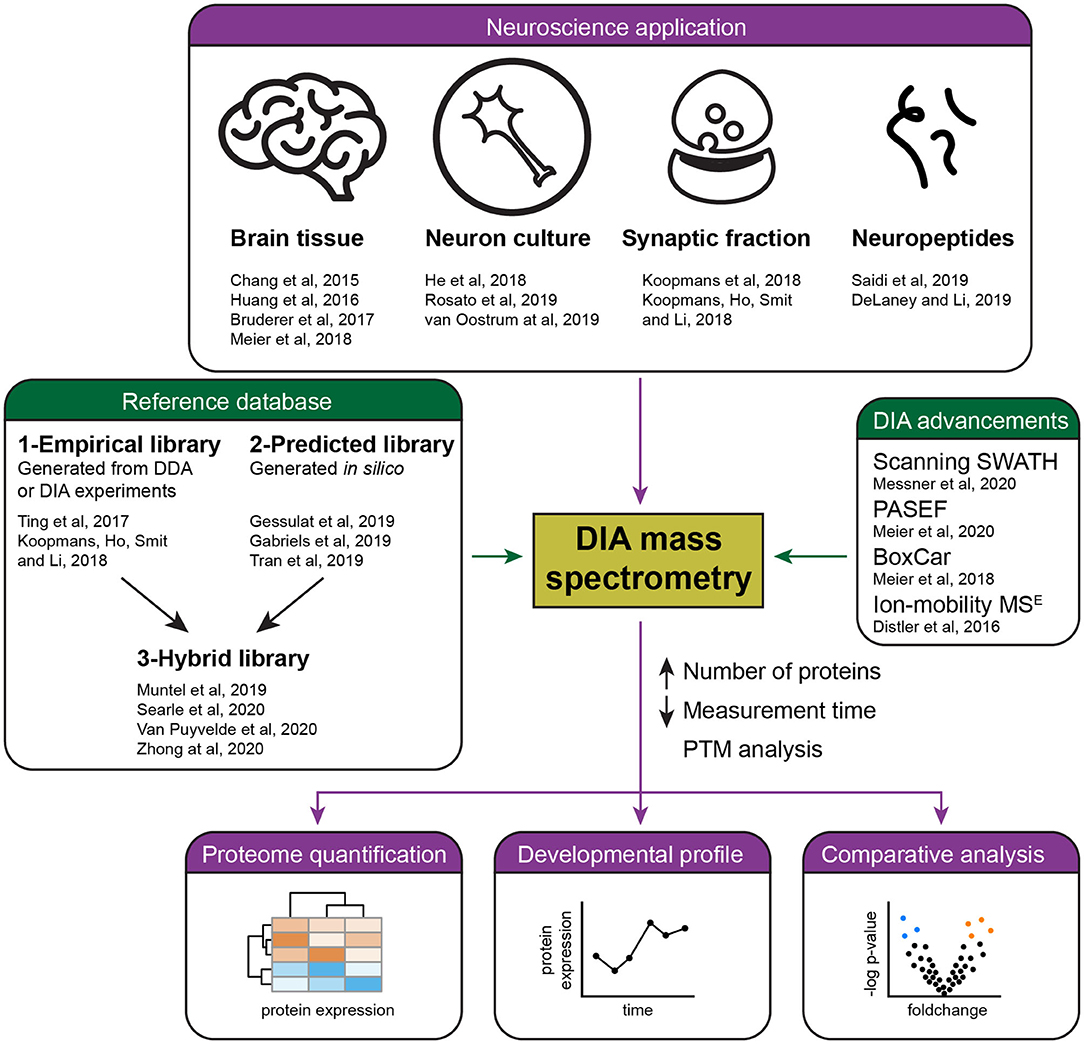
Frontiers | Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome

Hybrid Spectral Library Combining DIA-MS Data and a Targeted Virtual Library Substantially Deepens the Proteome Coverage - ScienceDirect
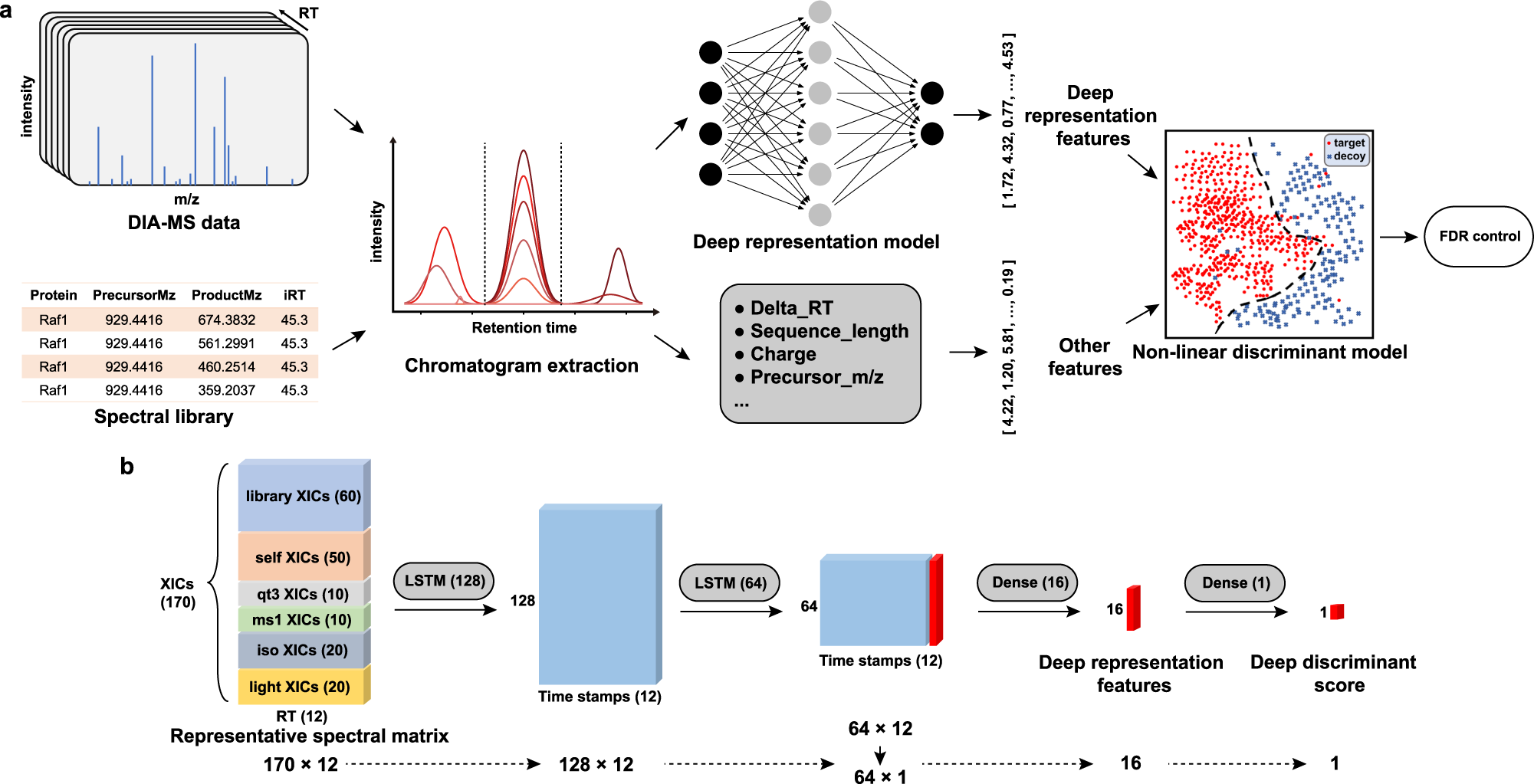
Deep representation features from DreamDIAXMBD improve the analysis of data-independent acquisition proteomics | Communications Biology
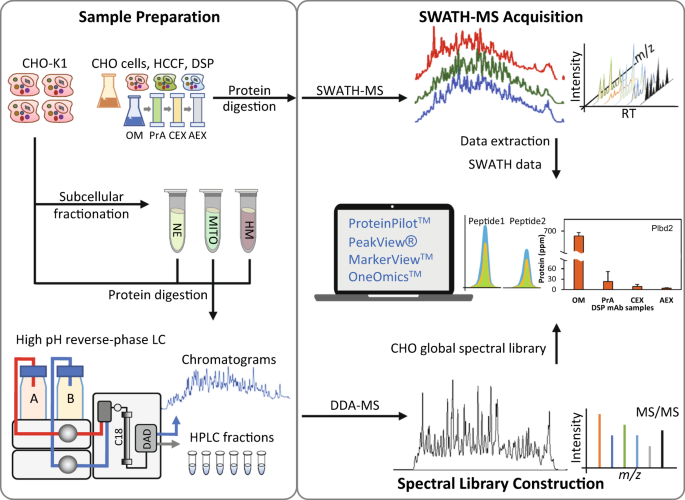
A comprehensive CHO SWATH-MS spectral library for robust quantitative profiling of 10,000 proteins | Scientific Data
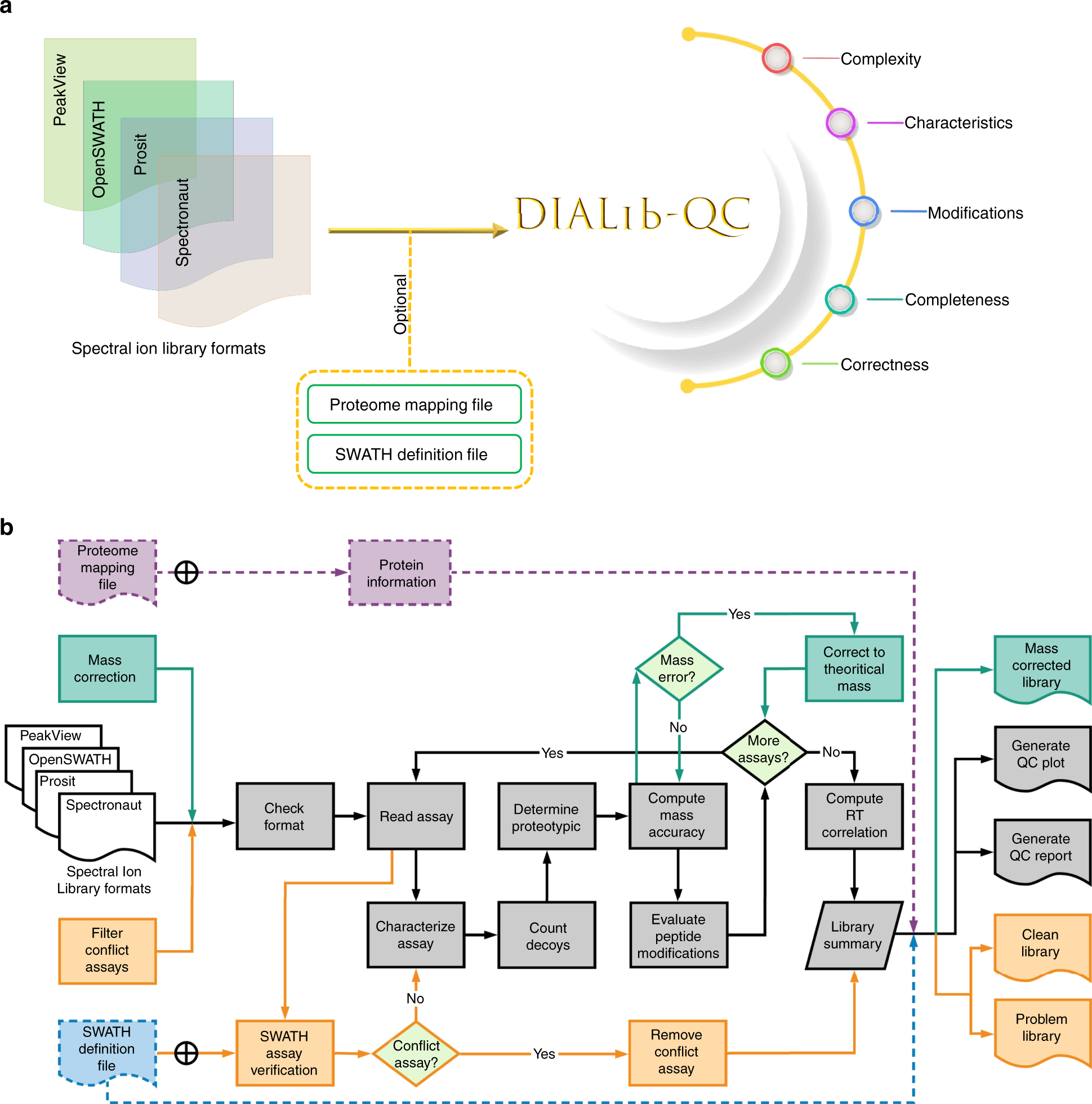
DIALib-QC an assessment tool for spectral libraries in data-independent acquisition proteomics | Nature Communications

Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications

Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files | Nature Methods
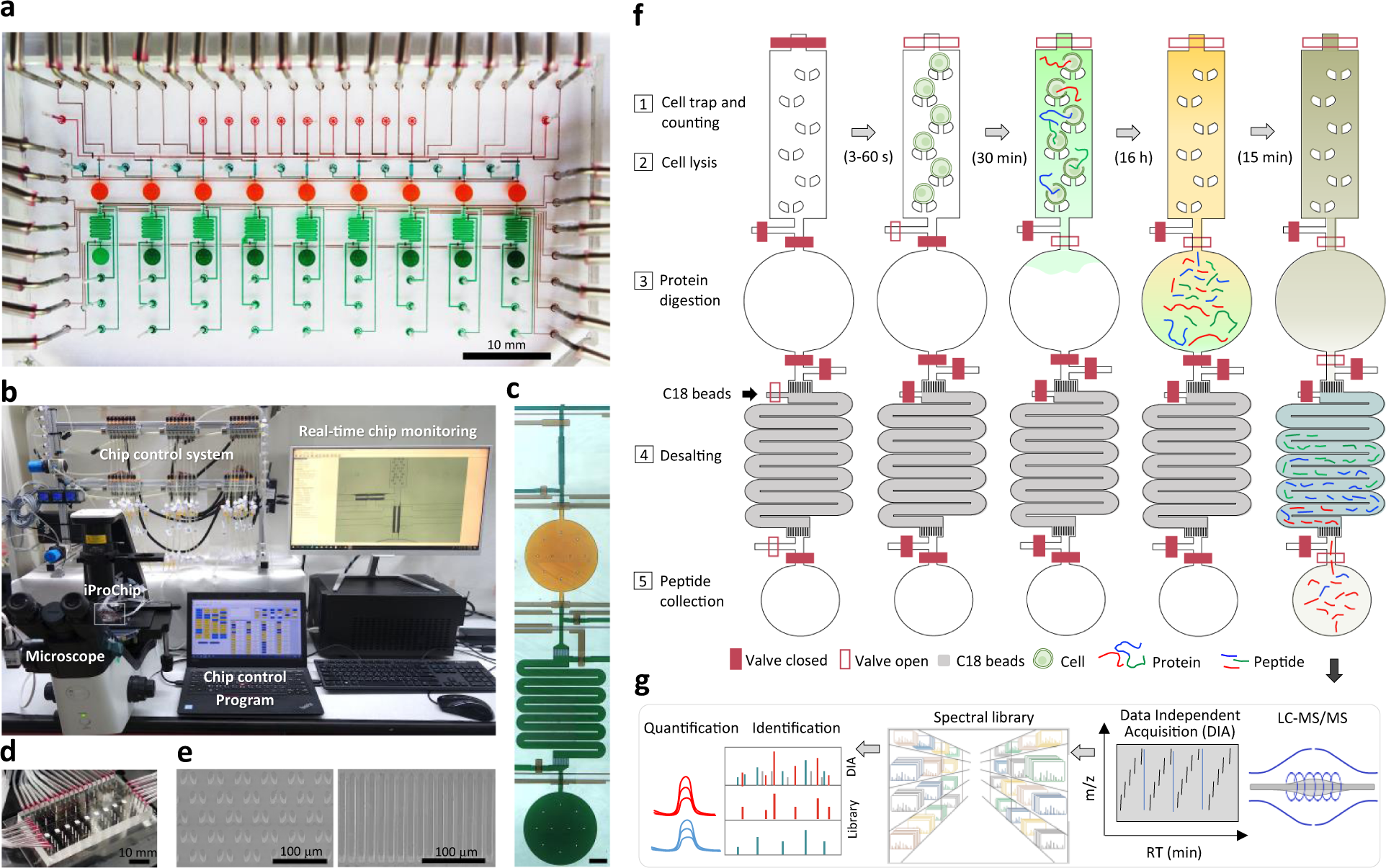
Streamlined single-cell proteomics by an integrated microfluidic chip and data-independent acquisition mass spectrometry | Nature Communications
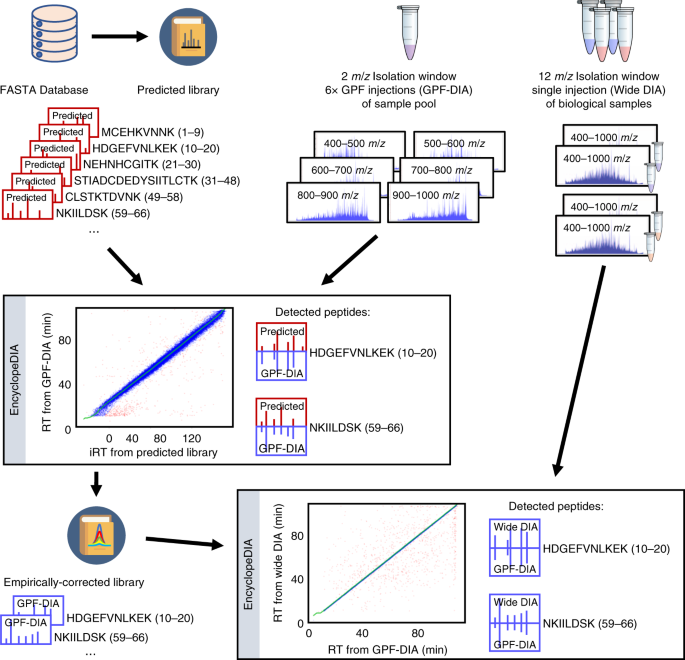
Generating high quality libraries for DIA MS with empirically corrected peptide predictions | Nature Communications
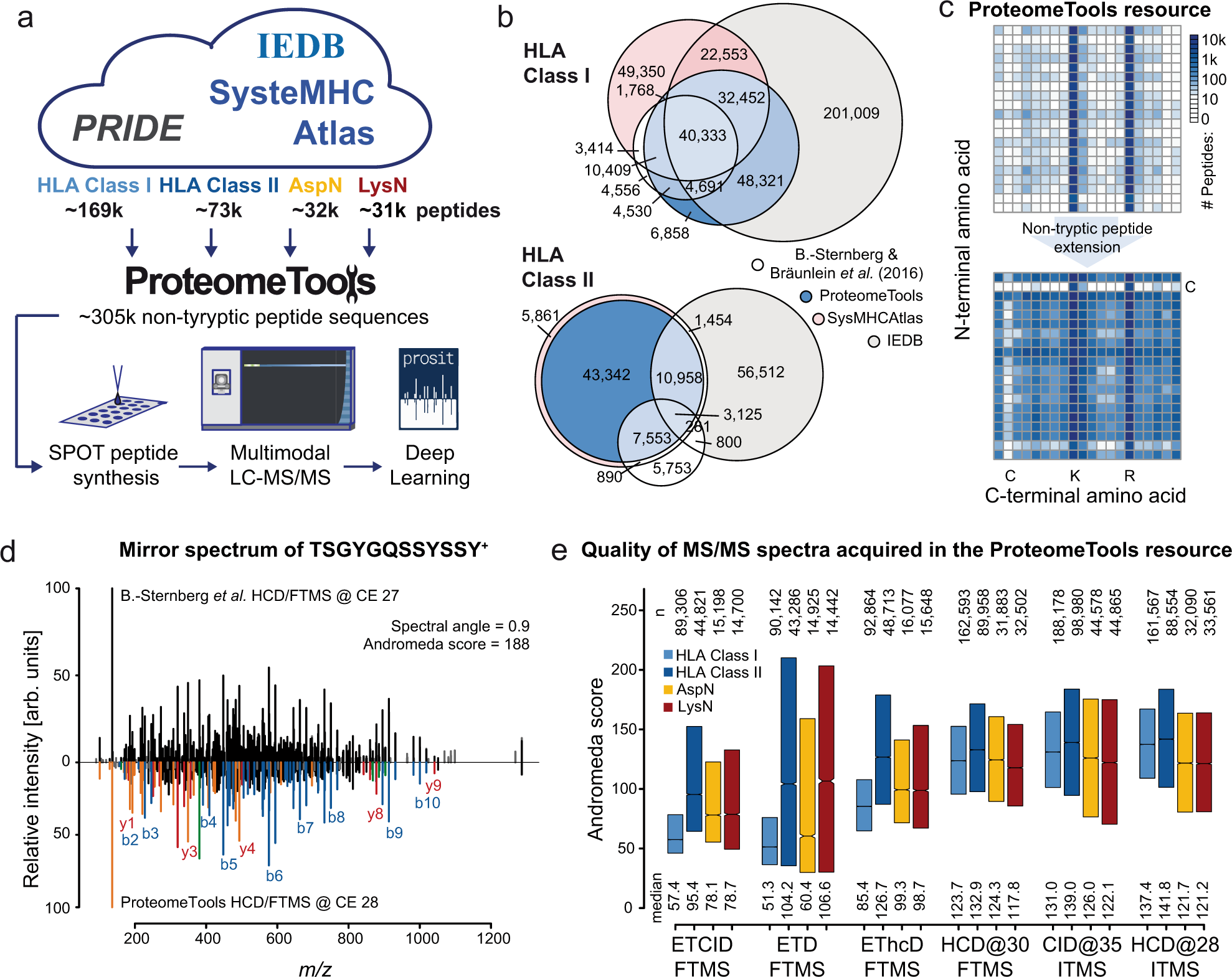
Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics | Nature Communications
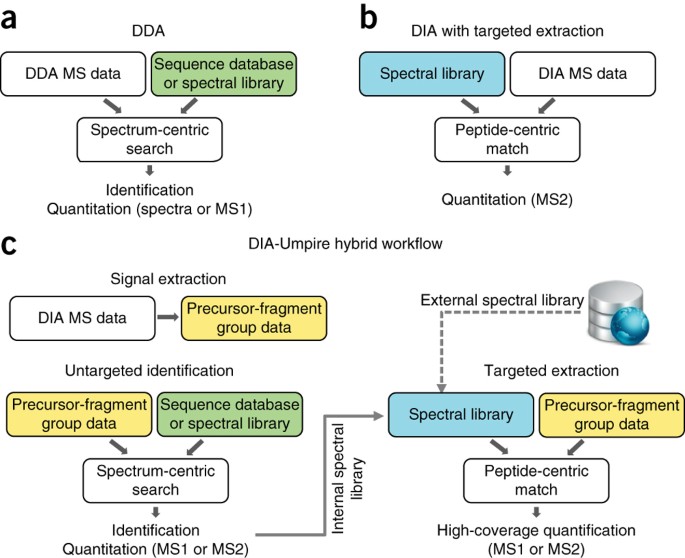
DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics | Nature Methods